A case for optimism in the age of AI
AlphaFold, Nobel Prizes, and a world where AI makes us smarter, healthier, and happier
Last month, Dario Amodei, CEO of Anthropic, wrote a piece titled “Machines of Loving Grace”. The title is a nod to Richard Brautigan’s 1967 poem that envisions a world where “mammals and computers live together in mutually programming harmony like pure water touching clear sky”.
Amodei writes in his piece to contemplate “what a world with powerful AI might look like if everything goes right”. Amidst a throng of pessimists and skeptics, much of which is merited, there is a path to a world where AI’s impact yields tremendous upside.
Dario’s piece delightfully coincided with the announcement of the Nobel Prize for Chemistry being awarded to David Baker of Baker Labs, and Demis Hassabis and John Jumper of Google DeepMind, for their groundbreaking research on protein structure, heavily enabled by AI.
Today, I’m going to discuss AlphaFold - what is does and why it matters - and a rose-colored perspective on a future where humans and AI can benefit tremendously from the speed of progress. We are just at the precipice, and we’re exploring this juncture through one of biology’s toughest problems.
The protein folding problem
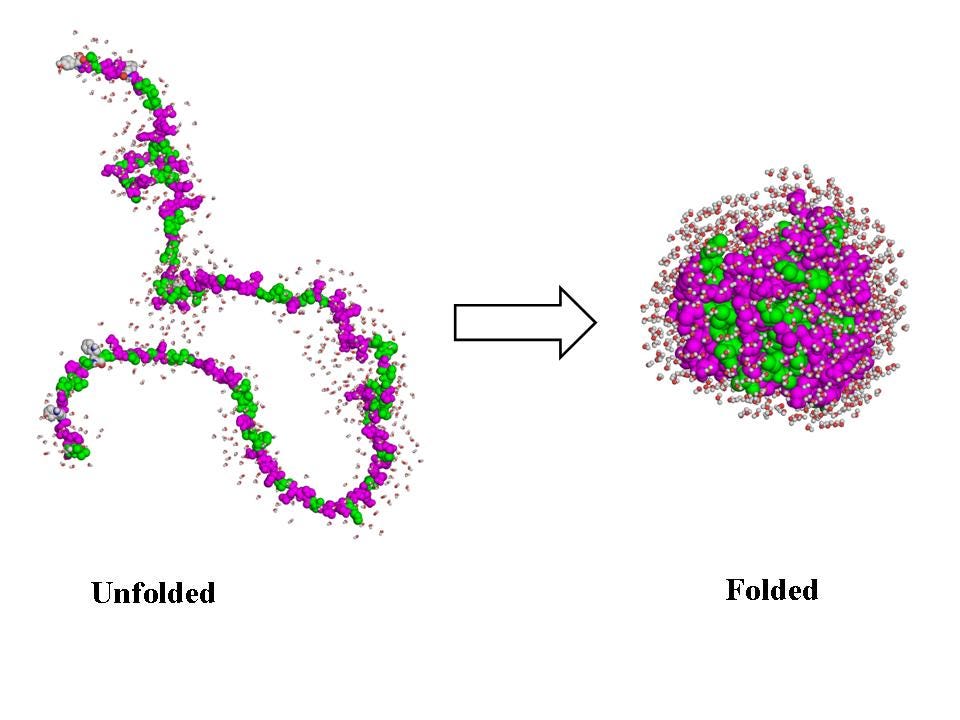
Proteins have been around for an estimated 3.7B years. Proteins start as a string of amino acids, which fold into a precise shape that enables specific functionality. The possible folding patterns are nearly infinite; Levinthal’s paradox suggests that testing every configuration by brute force would take longer than the age of the universe.
Understanding protein structures and their folding process can help scientists debunk protein behavior and function. This knowledge can get us closer to curing diseases that are results of anomalies in the folding process, like Alzheimer’s or Sickle Cell disease, as well as creating target drugs that work with a protein’s shape.
To understand the shape of a single protein, it could take hundreds of thousands of dollars and years of a researcher’s time - but what if it didn’t?
CASP & some healthy competition
There are three questions that scientists have wanted to understand about proteins that encompass the “protein folding problem”:
What is the folding code?
What is the folding mechanism?
Can we predict a protein’s structure, solely on its amino acid sequence?
In 1994, Professor John Moult and Professor Krzysztof Fidelis founded the Critical Assessment of Protein Structure Prediction (CASP) to inspire research teams every two years to test their protein folding prediction methodology.
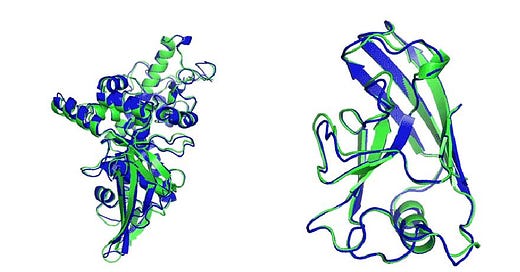
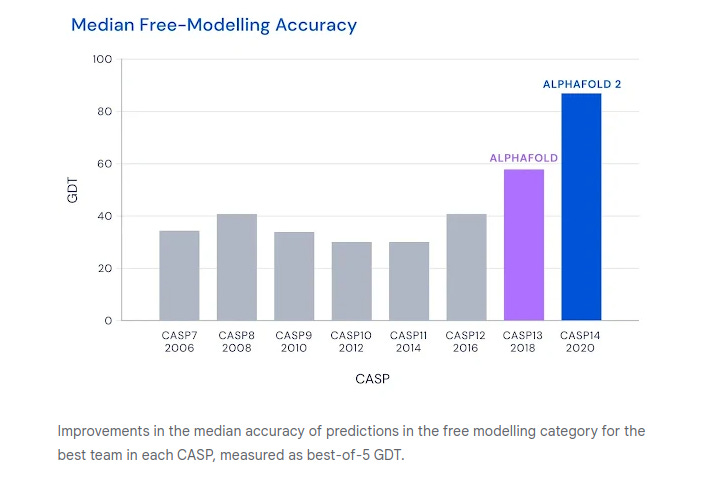
These methodologies are scored using the Global Distance Test (GDT)— the measure of similarity between two protein structures. In 2018, a brilliant team from Google’s DeepMind entered the 13th CASP competition, and their AlphaFold algorithm ended up beating the previous competition’s winning algorithm by almost 50%. It was an unprecedented outcome.
“We were the best team in the world at a problem the world was not very good at” — John Jumper, CASP 13 winning team member and Director at Google DeepMind
At the following competition in 2020, AlphaFold 2 achieved another record breaking score of 87.0 GDT. AlphaFold was no longer just a computer experiment, but a leap towards better medicine, healthier patients, next-gen biology, and an improved understanding of our world.
How AlphaFold works
AlphaFold follows two other impressive algorithms released by the Google team — AlphaGo and AlphaZero. AlphaGo most famously was an algorithm taught to play Go, a highly complex, strategic game that takes place on a 19x19 board. In 2016, AlphaGo beat Lee Sedol, a worth renowned Go player, in an incredible moment for the DeepMind team.

Biology, and the rules of physics at its core, is pretty beautiful in my opinion. The data about the world around us is full of patterns that are hard to interpret to the human eye, but that computers and algorithms can uncover across massive amounts of data.
It’s clear that AlphaFold 2 is learning something implicit about the structure of chemistry and physics. It sort of knows what things might be plausible. It’s learned that through seeing real protein structures, the ones that we know of. But one of the innovations we had was to do something called self-distillation, which is to get an early version of AlphaFold 2 to predict lots of structures—and to predict the confidence level in those predictions.— Demis Hassabis, CEO of DeepMind
From a one dimensional sequence of amino acids, AlphaFold predicts what the 3D protein shape is going to be, using solely machine learning methods, versus expensive and time consuming methods like X-ray crystallography or magnetic resonance spectroscopy.
AlphaFold was trained on the Protein Data Bank. At a very high level…. I’ll do my best to walk you through it. Using neural networks, the model takes an amino acid sequence as an input, and uses that to search against comparable proteins. Those comparable proteins and structural protein data are fed into an architecture called an EvoFormer, purpose built for this protein folding problem, which is then input to a structure module that predicts 3d atomic coordinates. The model then refines itself, taking one predicted structure and using it as input in the next iteration of prediction, helping the network get smarter with each turn. The output of many turns of this cycle is a set of predicted 3d atomic coordinates for resulting protein from a given amino acid sequence. Here’s a great video with a more technical breakdown of how the AlphaFold algorithm works.
The future with AlphaFold
AlphaFold left a remarkable impression on the research community. They’ve also since released further improved models, AlphaFold 2 and AlphaFold 3.
“AlphaFold changed the game,” Guo said. “In this [2022] competition, almost all groups used AlphaFold as a key component of their systems to make their predictions.” — CASP 15 winning team member, Zhiye Guo
AlphaFold’s impact doesn’t stop at medicine. By understanding protein folding, we can tackle problems like:
Helping fight malaria, that can improve the efficacy of the malaria vaccine
Supporting work towards a plastic polluting enzyme
Predicting the structure of PINK1, a protein that impacts those with Parkinson’s
Getting closer to solving these questions, and the hundreds of other that AlphaFold has gotten us a bit closer to cracking, is a world I am excited to live in.
The “compressed 21st century”
Dario Amodei coined a phrase that really stuck with me in his piece — the “compressed 21st century”.
My basic prediction is that AI-enabled biology and medicine will allow us to compress the progress that human biologists would have achieved over the next 50-100 years into 5-10 years. I’ll refer to this as the “compressed 21st century”: the idea that after powerful AI is developed, we will in a few years make all the progress in biology and medicine that we would have made in the whole 21st century. — Dario Amodei
It’s a good reminder of the value in investing in resilient and adaptable companies, capable of being improved by rapid technological advancement.
We are standing on the brink of a compressed century—a period that might condense decades of progress into just a few years. Like the leap from steam engines to electricity, which took 100+ years, the speed of AI suggests we’re in for a massive acceleration of progress. We are just getting started.
Thanks for reading October’s Day to Data (though a few days late!). October was a busy month, full of running the Chicago Marathon, enjoying an unseasonably warm NYC fall, and cooking new meals at home! The next few months include a trip to Atlanta for Supercomputing and Vancouver for NeurIPS - if you’ll be in town for either, shoot me a message. See you all later this month.



Incredible. My head spins just thinking of all the ideas captured here. I am agnostic as to the value of AI, given that it can be used with malice by parties with bad intentions and it will be hard to predict a potential lethal, game-changing path from which humanity may not be able to recover. That said, the application of AI to the natural sciences gives me reason for optimism.